[By Aaron Dubrow, originally published on the TACC website]
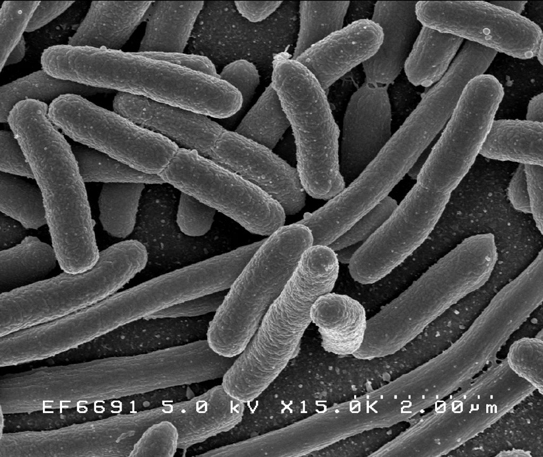 Escherichia coli: Scanning electron micrograph of Escherichia coli, grown in culture and adhered to a cover slip. [Credit: Rocky Mountain Laboratories, NIAID, NIH]
Escherichia coli: Scanning electron micrograph of Escherichia coli, grown in culture and adhered to a cover slip. [Credit: Rocky Mountain Laboratories, NIAID, NIH]
E. coli bacteria multiplied in their Erlenmeyer flasks, evolving slowly over more than 50,000 generations. Throughout that time, scientists at Michigan State University kept records of each successive generation, waiting for the telltale signs of evolution to show themselves.
The most dramatic sign first appeared after approximately 33,000 generations in the form of a cloudy bloom of bacteria in one flask.
"Over time, the E. coli developed a mutation that allowed them to have a leap in function, an innovation," said Jeffrey Barrick, a former researcher in the Michigan State lab, and now professor of chemistry and biochemistry at The University of Texas at Austin. "It was feeding on citrate, an untapped carbon source that had been there all along. This enabled the bacteria that mutated to have a huge advantage, to take over the whole population, and even grow to a higher population density inside of this flask, which you could see by eye."
Typically, the inability of E. coli to consume citrate in normal conditions is a defining feature of the species. How did it suddenly overcome its deep-rooted bias?
Based on the recordings of each successive generation, the researchers identified not only the final cells that had realized this new potential, but also all the preceding lines whose subtle, latent changes had enabled the final ability to emerge.
"About 70 or 80 changes occurred between the ancestor and this final bacterium," Barrick said. "We picked a bunch of individuals from the population at different time points and created a phylogenetic tree." As part of his research, Barrick models the ways that emerging mutations flow through populations. Some evolutionary changes radically alter an organism's ability to compete; some never catch on. The difference is a function of luck as well as the evolutionary advantages of a given mutation and how mutations interact with one another. Understanding the interplay between these factors could help researchers create useful organisms through smart evolution. Credit: Jeffrey Barrick, Austin Meyer, The University of Texas at Austin.
As part of his research, Barrick models the ways that emerging mutations flow through populations. Some evolutionary changes radically alter an organism's ability to compete; some never catch on. The difference is a function of luck as well as the evolutionary advantages of a given mutation and how mutations interact with one another. Understanding the interplay between these factors could help researchers create useful organisms through smart evolution. Credit: Jeffrey Barrick, Austin Meyer, The University of Texas at Austin.
Using next-generation DNA sequencers and the powerful Lonestar and Ranger supercomputers at the Texas Advanced Computing Center (TACC) to test 40 genomes from the population, the researchers traced the key changes that potentiated the mutation and showed the role of promoter capture and altered gene regulation in evolutionary innovations.
The results of the study were published in Nature in September 2012.
To make this discovery, Barrick used a host of technologies that have made DNA sequencing cheaper, faster, and more accurate in recent years. He also developed tools, including breseq, which are capable of finding more, and more difficult-to-locate, mutations.
"None of the other tools were up to par," he explained. "They didn't find these other categories of mutations. And from other evidence, we knew they weren't finding about one-third of the mutations that were happening."
Duplications, reordering, and mobile genetic elements that can jump to different parts of the genome are hard to identify with conventional tools, but are important for the evolution of novelty because they change the genome more than one letter at a time, sometimes rearranging sequences entirely.
"Breseq is a transformative application for our field," said Vaughn Cooper, an associate professor of molecular, cellular and biomedical sciences at the University of New Hampshire. "It has made what was typically a month-long process of data filtering and analysis possible within a day or two, even without high-power computing. Supercomputing would make the output from his tool applied to even larger data sets virtually instantaneous."
If the E. coli experiment showed the potential of genetic analysis to reveal the path to evolutionary innovation and to map them across generations, the work that interests Barrick now applies this understanding to the design of artificial life.
"I think a lot about the engineering aspect: making bacteria do useful things," Barrick said. "I would like bacteria to solve our energy crisis, whether that means making biofuels or something crazy like putting molecular motors in algae that push water to run a generator."
Despite all we know about E. coli, scientists are hard pressed to pick mutations that make bacteria really good at a given process. But Barrick believes that understanding where to make those changes, and how to make different kinds of changes, could make the process of evolutionary design smarter and more efficient.
His report on efforts to use computer algorithms to test strategies for improving the evolution of complex functional nucleic acids was awarded Best Synthetic Biology Paper at Artificial Life XIII: Proceedings of the Thirteenth International Conference on the Synthesis and Simulation of Living Systems in July 2012.
Across the field, advanced computing is allowing researchers like Barrick to analyze genomes, develop and test synthetic organisms, and experiment with artificial populations, all of which help scientists explore evolution on a far finer-grained level.
"When this E. coli experiment started, all they could measure was fitness. They had no clue why certain E. coli strains were better," Barrick said. "Now we can understand at the molecular level what's going on, and that's really powerful."

















Comments 1
Let's be honest.
Nobel Laureate WERNER ARBER, a Swiss micro-biologist, has spent the last 51 years of his life, documenting the cumulative effects of MUTATIONS on thousands upon thousands of generations of single-cell organisms. Thus far, Werner Arber reports that he has found ... NO MECHANISM ... that would enable a single-cell organism to evolve into a multi-cell organism.